Contrasting diversity of phycodnavirus signature genes in two large and deep western European lakes
Abstract
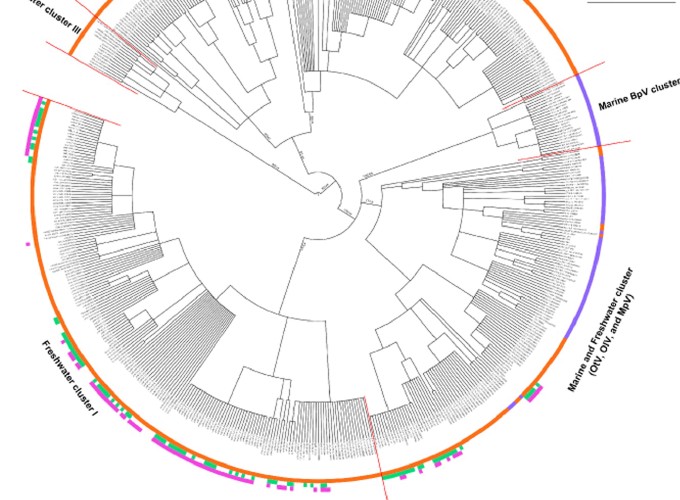
Little is known about Phycodnavirus (or double‐stranded DNA algal virus) diversity in aquatic ecosystems, and virtually, no information has been provided for European lakes. We therefore conducted a 1‐year survey of the surface waters of France’s two largest lakes, Annecy and Bourget, which are characterized by different trophic states and phytoplanktonic communities. We found complementary and contrasting diversity of phycodnavirus in the lakes based on two genetic markers, the B family DNA polymerase‐encoding gene (polB) and the major capsid protein‐encoding gene (mcp). These two core genes have already been used, albeit separately, to infer phylogenetic relationships and genetic diversity among members of the phycodnavirus family and to determine the occurrence and diversity of these genes in natural viral communities. While polB yielded prasinovirus‐like sequences, the mcp primers yielded sequences for prasinoviruses, chloroviruses, prymnesioviruses and other groups not known from available databases. There was no significant difference in phycodnavirus populations between the two lakes when the sequences were pooled over the full year of investigation. By comparing Lakes Annecy and Bourget with data for other aquatic environments around the world, we show that these alpine lakes are clearly distinct from both other freshwater ecosystems (lakes and rivers) and marine environments, suggesting the influence of unique biogeographic factors.
More detail can easily be written here using Markdown and $\rm \LaTeX$ math code.